1587
Views & Citations587
Likes & Shares
The effectiveness of
drug combinations for treatment of a variety of complex diseases is well
established. “Drug cocktail” treatments are often prescribed to improve the
overall efficacy, decrease toxicity, alter pharmacodynamics, etc., in an
overall treatment strategy. Specifically, if when combined, drugs interact in
some way that causes the total effect to be greater than that predicted by
their individual potencies, then drugs are considered synergistic. While there
are established ways to quantify the impact of drug combinations clinically, it
is an open challenge to quantitatively summarize a synergistic interaction. In
this paper, we discuss an overview of the current statistical and mathematical
methods for the study of drug combination effects, especially drug synergy
quantification (where the interaction effect is not just detected, but
quantified according to its magnitude). We first introduce two popular
reference models for testing to null hypothesis of non-interaction for a
combination, including the Bliss independence model and the Loewe additivity
model. Then we discuss several methods for quantifying drug synergism. The
advantages and disadvantages with these methods are also provided, and finally,
we discuss important next directions in this area.
INTRODUCTION
For
a variety of complex diseases, it is an accepted paradigm that drugs are given
in combination [1]. A drug interaction is a situation in which another drug
affects the activity of a drug when both are administered together. This action
can be synergistic (when the drug's effect is increased) or antagonistic (when
the drug's effect is decreased) [2]. The evaluation of combination effects
between biological or chemical agents plays a significant role in pharmacology
and biomedicine. Combination therapies, often referred to as “cocktail”
therapies have revolutionized patient outcomes in diseases such as HIV [3],
asthma [4], breast cancer [5,6], hypertension [7] and cancers such as melanoma
[8]. The impact of chemical mixtures is also increasingly appreciated in the
toxicology space as well, as people are not exposed to chemicals in isolation
[9]. A recent review discusses the concept of synergy as used in a variety of
fields [10].
The
interaction of biologically or chemically active agents is often grouped into
three categories: synergy, additivity (no interaction) and antagonism, based on
the degree of departure of observed combination effects from the expected response
without interaction [2,11]. Specifically, if drugs when combined interact with
each other and cause a total effect that is greater than that predicted by
their individual potencies, then this is considered a “synergistic drug”
combination [12]. Such synergistic interactions can often reduce host toxicity
and adverse side effects, given those doses of each drug in the combination are
typically lower than that of single drugs to achieve desired efficacy.
Additionally, such combination therapies can also reduce the development of
drug resistance and other complications [13,14].
In
this review, we provide an overview of the current statistical and mathematical
methods for the study of drug combination effects, especially drug synergy quantification.
We first introduce two popular reference models for the null hypothesis of
non-interaction, which serve as the baseline to define synergy. Any deviation
from the reference models will be regarded as synergy or antagonism.
Subsequently, we discuss several statistical and mathematical approaches to
quantify drug synergism. Finally, the common issues and opportunities with
these methods are also provided. Although this paper mainly covers drug
synergy, the concepts and methods mentioned in this review can be applied to
other disciplines as well, such as toxicology and epidemiology.
TWO REFERENCE MODELS
To properly define synergy, it is of great
importance to formulate a reference model for null hypothesis of
non-interaction first, which suggests that the effects of drugs simply add up,
not affecting each other (Additivity) [15]. Any deviation from the reference
models will be regarded as synergy or antagonism, depending on the directions
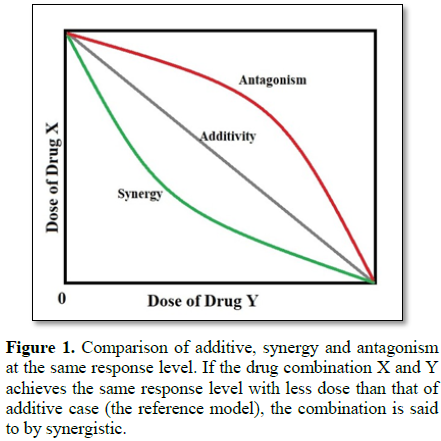
of departure. As shown in Figure 1,
if the drug combination X and Y achieves the same response level with less dose
than that of additive case (the reference model), the combination is said to by
synergistic. Currently, there are two popular reference additivity models,
Bliss independence model and the Loewe additivity model, which have different
biological and chemical assumptions.
Bliss independence model
One
of the oldest methods for quantifying synergy is the Bliss Independence model,
dating back to the 1930s [16]. This model assumes that drugs do not interact with
each other and elicit their responses independently [1]. According to
independence probability theory, the expected response of drug combination Rc
(0 ≤ Rc ≤ 1) can be written in terms of individual drug responses
[16]:
Rc = R1 + (1 - R1)
× R2 = R1 + R2 - R1 × R2
Where
drug 1 at dose y1 produced a response R1, drug 2 at dose
y2 produced a response R2 and Rc is the
expected response of drug combination 1 and 2 at dose y1 and y2,
respectively. As the drug’s effects R1, R2 and Rc
are measured as the percentage of biological response, 0 ≤ R1 ≤ 1, 0
≤ R2 ≤ 1 and 0 ≤ Rc ≤ 1. Any observed response of drug
combination greater than the expected response Rc can be interpreted
as synergy and antagonism otherwise.
This
null model is classically known in toxicology as “simple independent action”
and is based on probabilistic independence. The paradigm is where there are two
disjoint and independent causal pathways on which the two drugs act. The above
equation can alternatively simply be rewritten as additivity in the logarithms
of the two probabilities of nonresponse.
Loewe additivity model
An alternative null model is the additivity model,
which assumes that drugs have similar modes of action on the same pathway [1,17].
In classical toxicology, this model is known as “simple similar action.” It
specifies that one drug’s dose has the same effect on response as a scaling
factor times the other drug’s dose. To formulate this as specified in the Loewe
additivity model, the dose-response relationship of individual drugs needs to be
modeled first. Let the dose of drug 1=y1 and the dose of drug 2=y2.
Then the Loewe additivity model can be expressed as the following equation [1,17]:
(y1 / Y1) + (y2
/ Y2) = 1
Where
Y1 is the dose of drug 1 that achieves the same response level as
the drug combination, y1+y2 and Y2 is the dose
of drug 2 that achieve the same response level as the drug combination. The
left side of this equation is the widely used combination index. If a
combination index is less than 1, synergy is declared. Similarly, a combination
index greater than 1 can be interpreted as antagonism.
The
major differences of the two reference models come from their underlying
assumptions. The Bliss independence model assumes that drugs do not interact
with each other and elicit their responses independently, whereas the Loewe
additivity model assumes that drugs have similar modes of action on the same
pathway. Note that if the response is rare, the two formulations are asymptotically
equivalent. In practice, the selection of one of those two to serve as the null
model for assessing synergy and antagonism becomes largely a matter of personal
preference [18]. To address the concerns raised in the two reference models, Yadav
et al. [19] recently proposed a new reference model called zero interaction
potency (ZIP). It evaluates drug interaction by comparing the change in dose
response relationships between single drugs and their combinations. The results
show that this new scoring method is able to keep the advantages of the two
popular reference models mentioned above while overcome their limitations.
THE METHODS FOR QUANTIFYING DRUG SYNERGISM
Next,
we will discuss methods for actually directly quantifying synergy. The methods
are briefly introduced here, with references provided for a more detailed
description of each approach.
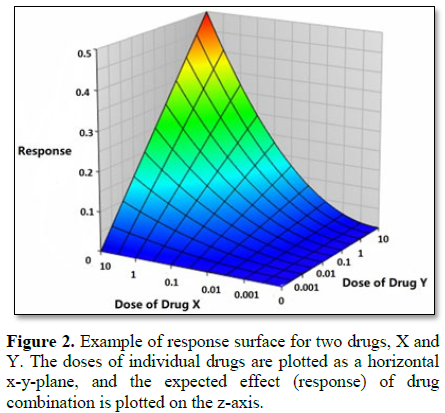
Response surface
Response
surface modeling is an approach to represent effects of drug combinations in
three-dimensional plots where the doses of individual drugs are plotted as a
horizontal x-y-plane, and the expected effect of drug combination is plotted on
the z-axis, as shown in Figure 2
[1]. Both the Bliss independence model and the Loewe additivity model can be
used to calculate the expected effect of drug combination [1]. The experimental
effect of a drug combination can then be plotted on this surface. Any departure
from the 3D null surface is classified into synergism or antagonism, depending
on the sign of the discrepancy as measured on the z-axis. [20,21].
Chou-Talalay method
The Chou-Talalay method is by far the most commonly
used approach to quantify effects of drug combinations, especially synergistic
interactions [15,21]. This method adopts the median-effect equation, which is
derived from the unified theory mass-action law principle [9]. The
median-effect equation is written below [21]:
fa / fu = (D / Dm)m
Where
fa is the fraction affected by dose, fu is the fraction
unaffected by dose (fa+fu=1), D is dose of drugs given, Dm
is the median-effect dose (e.g. IC50) and m is a parameter used to describe the
shape of the dose-response curve.
The
median-effect equation can be linearized by taking logarithm of both sides of
the equation, as shown below:
Log
(fa / fu) = m × log (D) – m × log (Dm)m
Then
the values of m and Dm can be estimated using linear regression.
With this linear model, we can estimate the expected drug response values given
specific drug doses, which will be used in the calculation of combination index
(CI):
CI = D1 / Dx1 + D2
/ Dx2
Where
D1 and D2 are the doses of two single drugs and Dx1
and Dx2 is the theoretical individual drug doses needed in order to
achieve the same drug effect as the drug combination, which can be calculated
based on the linear model mentioned above. CI<1 suggests synergism, CI=1
suggests additivity and CI>1 suggests antagonism [15,22].
One
disadvantage of the Chou-Talalay method is that raw data must be preprocessed,
including scaling the data and taking the log of a function of the scaled data
[15].
MixLow method
More
recently, Boik et al. [23] developed the MixLow method as an alternative to the
Chou-Talalay method. The term MixLow means Mixed-effects Loewe, which has three
components: a nonlinear mixed-effects model, the Loewe index and a method to
calculate confidence intervals for the index. The MixLow method uses a
nonlinear mixed effects model for estimating sigmoidal curve parameters from
concentration-response data, and associated confidence intervals [23]. Compared
with the Chou-Talalay method, the MixLow method produces more precise parameter
estimation, and has improved coverage of confidence intervals. In addition, the
use of a non-linear fixed-effects model in the MixLow method also eliminates
the need for data preprocessing in the Chou-Talalay method [23].
Drug synergy quantification using a Bayesian
approach
In
2010, Hennessey et al. proposed a Bayesian approach to dose-response assessment
and synergy quantification. Briefly, they use a Bayesian hierarchical nonlinear
regression model to explain the “variability between-experiments, variability within
experiments, and variability in the observed responses of the controls” [24].
They first use Markov chain Monte Carlo (MCMC) to fit the model to the data.
The second step is to carry out posterior inference on quantities of interest.
Finally, they assess the presence of synergy while accounting for uncertainty
using a modified version of Loewe additivity. Simulation results suggest that
this method is more reliable in drug synergy estimation than the Chou-Talalay
method, which often ignores important sources of variability and uncertainty
that is generally the rule, instead of the exception in biology [24].
Summary
of advantages and disadvantages of current methods (Table 1)
CURRENT PROBLEMS AND
FUTURE DEVELOPMENTS IN DRUG SYNERGY QUANTIFICATION
Drug
combinations provide many advantages in the treatment of complex disease. The
search for drug combinations has been widely recognized as one of the most
important strategies for finding successful treatments of cancer and other
diseases [15]. Although recent methods development has improved, there are
still a number of open challenges and issues that need to be addressed.
First,
there are still a number of challenges related to even defining synergy, much
less quantifying it. In the current literature, the term synergy is not often
clearly defined. Research papers usually use different reference models to
quantify synergy in particular cases, which causes lack of comparability and
confusion [18,25]. Thus, a standard reference framework should be developed to
address the concerns raised in the current reference models and provide a clear
definition of additivity, synergy and antagonism. Additionally, the standard
framework should also be general enough to cover rare and specific cases so
that researchers can use a universal method to quantify drug synergy. Our group
has recently reviewed some of the challenges and differences in the terminology
related to synergy [10].
Additionally,
there are outstanding challenges in experimental design that need to be
considered and advanced. One of the most important challenges of any study that
will study synergy is the selection of dose and dose ratios in drug combination
studies. The advantages of combination therapy not only depends on the
properties of the drugs but also depend on the dose ratios [26,27]. Considering
that two drugs combined at a given ratio are often treated as a new drug with
its own dose-effect relationship in cells and tissues, we not only need to
study whether a particular combination is synergistic, we also need to consider
what dose ratio optimizes the synergistic interaction [26]. This is important
in both experimental studies, and in clinical application.
Finally,
we need to keep advancing more rigorous statistical methodology to interpret
the variation in drug synergy quantification. Current methods quantify synergy,
but do not ascribe a statistical confidence level with those estimates. Data
from biological systems always carry experimental error and there is also
inherent biological variation. However, the most commonly used combination
indexes based on Bliss Independence and Loewe Additivity are often calculated
without a suitable error assessment to measure the degree of uncertainty. The
lack of a formal statistical framework in these approaches makes it difficult
to interpret drug combination effects, especially for borderline cases.
CONCLUSION
In
the current review, we discuss an overview of the current statistical and
mathematical methods for the study of drug combination effects, especially drug
synergy quantification. We introduce two popular reference models for
non-interaction of a combination, including the Bliss independence model and
the Loewe additivity model. Then we discuss several methods for quantifying
drug synergism. The advantages and disadvantages associated with these methods
are also provided, and finally, we discuss current problems and future
developments in drug synergy quantification.
Addressing
these limitations represents an important methodological research direction. Recently
there have been a number of new approaches to quantify dose response curves
using machine learning methods, including evolutionary algorithms [28]. Such an
approach could be extended to the drug combination effects as well.
Advances in the
statistical methods will allow researchers to estimate the
variability in biological
or clinical experiments with sufficient accuracy and further improve the degree
of confidence in drug synergy detection. Moreover, these advances will also
benefit high-throughput drug combination screening greatly. The integration of
automated screening techniques with robust statistical methods will facilitate
the discovery of reliable synergistic drug interactions, ultimately improving
the sensitivity and specificity of the screening process. Although we mainly
discuss drug synergy here, these advances in statistical methods can be easily
applied to other disciplines as well, such as environmental toxicology and
epidemiology. For instance, we can detect the combination effects of multiple
environmental chemicals for risk assessment purposes with a high degree of
confidence.
ACKNOWLEDGEMENT
This
research was supported by the Intramural Research Program of the NIH, National
Institute of Environmental Health Sciences.
1.
Greco WR, Bravo G, Parsons JC (1995) The search for
synergy: A critical review from a response surface perspective. Pharmacol Rev
47: 331-385.
2.
Geary N (2013) Understanding synergy. Am J Physiol
Endocrinol Metab304: 237-253.
3.
Bartlett JA, DeMasi R, Quinn J, Moxham C, Rousseau F
(2001) Overview of the effectiveness of triple combination therapy in
antiretroviral-naive HIV-1 infected adults. AIDS 15: 1369-1377.
4.
Nelson HS (2001) Advair: Combination treatment with
fluticasone propionate/salmeterol in the treatment of asthma. J Allerg Clin
Immunol 107: 398-416.
5.
Budman DR, Calabro A, Rosen L, Lesser M (2012)
Identification of unique synergistic drug combinations associated with down
expression of survivin in a preclinical breast cancer model system. Anticancer
Drugs 23: 272-279.
6.
Lee JH, Nan A (2012) Combination drug delivery
approaches in metastatic breast cancer. J Drug Deliv 2012: 915375.
7.
Glass G (2004) Cardiovascular combinations. Nat Rev
Drug Discov 3: 731-732.
8.
Held MA, Langdon CG, Platt JT, Graham-Steed T, Liu Z,
et al. (2013) Genotype-selective combination therapies for melanoma identified
by high-throughput drug screening. Cancer Discov 3: 52-67.
9.
Carpenter DO, Arcaro K, Spink DC (2002) Understanding
the human health effects of chemical mixtures. Environ Health Perspect 110:
25-42.
10.
Roell KR, Reif DM, Motsinger-Reif AA (2017) An
introduction to terminology and methodology of chemical synergy-perspectives
from across disciplines. Front Pharmacol 8: 158.
11.
Berthoud HR (2013) Synergy: A concept in search of a
definition. Endocrinology 154: 3974-3977.
12.
Cedergreen N (2014) Quantifying synergy: A systematic
review of mixture toxicity studies within environmental toxicology. PLoS One 9:
e96580.
13.
Greco WR, Faessel H, Levasseur L (1996) The search for
cytotoxic synergy between anticancer agents: A case of Dorothy and the ruby
slippers? J Natl Cancer Inst 88: 699-700.
14.
Worthington RJ, Melander C (2013) Combination
approaches to combat multidrug-resistant bacteria. Trends Biotechnol 31:
177-184.
15.
Chou TC (2010) Drug combination studies and their synergy
quantification using the Chou-Talalay method. Cancer Res 70: 440-446.
16.
Bliss CI (1939) The toxicity of poisons applied
jointly. Ann Appl Biol 26: 585-615.
17.
Finney DJ, Uhbgcbie R (1964) Statistical method in
biological assay. Hafner Pub. Co.
18.
Tang J, Wennerberg K, Aittokallio T (2015) What is
synergy? The Saariselka agreement revisited. Front Pharmacol 6: 181.
19.
Yadav B, Wennerberg K, Aittokallio T, Tang J (2015)
Searching for drug synergy in complex dose-response landscapes using an
interaction potency model. Comput Struct Biotechnol J 13: 504-513.
20.
Prichard MN, Shipman C Jr. (1990) A three-dimensional
model to analyze drug-drug interactions. Antiviral Res 14: 181-205.
21.
Chou TC, Talalay P (1984) Quantitative analysis of
dose-effect relationships: The combined effects of multiple drugs or enzyme
inhibitors. Adv Enzyme Regul 22: 27-55.
22.
Chou TC (2006) Theoretical basis, experimental design
and computerized simulation of synergism and antagonism in drug combination
studies. Pharmacol Rev 58: 621-681.
23.
Boik JC, Newman RA, Boik RJ (2008) Quantifying
synergism/antagonism using non-linear mixed-effects modeling: A simulation
study. Stat Med 27: 1040-1061.
24.
Hennessey VG, Rosner GL, Bast RC Jr., Chen MY (2010) A
Bayesian approach to dose-response assessment and synergy and its application
to in vitro dose-response studies.
Biometrics 66: 1275-1283.
25.
Jia J, Zhu F, Ma X, Cao Z, Cao ZW, et al. (2009)
Mechanisms of drug combinations: Interaction and network perspectives. Nat Rev
Drug Discov 8: 111-128.
26.
Keith CT, Borisy AA, Stockwell BR (2005) Multicomponent
therapeutics for networked systems. Nat Rev Drug Discov 4: 71-78.
27.
Straetemans R, O'Brien T, Wouters L, Van Dun J, Janicot
M, et al. (2005) Design and analysis of drug combination experiments. Biom J
47: 299-308.
28.
Beam AL, Motsinger-Reif AA (2011) Optimization of
nonlinear dose- and concentration-response models utilizing evolutionary
computation. Dose Response 9: 387-409.
QUICK LINKS
- SUBMIT MANUSCRIPT
- RECOMMEND THE JOURNAL
-
SUBSCRIBE FOR ALERTS
RELATED JOURNALS
- Advances in Nanomedicine and Nanotechnology Research (ISSN: 2688-5476)
- Journal of Agriculture and Forest Meteorology Research (ISSN:2642-0449)
- Journal of Genomic Medicine and Pharmacogenomics (ISSN:2474-4670)
- Journal of Genetics and Cell Biology (ISSN:2639-3360)
- Journal of Womens Health and Safety Research (ISSN:2577-1388)
- Journal of Microbiology and Microbial Infections (ISSN: 2689-7660)
- Journal of Biochemistry and Molecular Medicine (ISSN:2641-6948)